|
|
TASSEL Version 5.0
(Getting Started!)
Tassel 5 Mac OS |
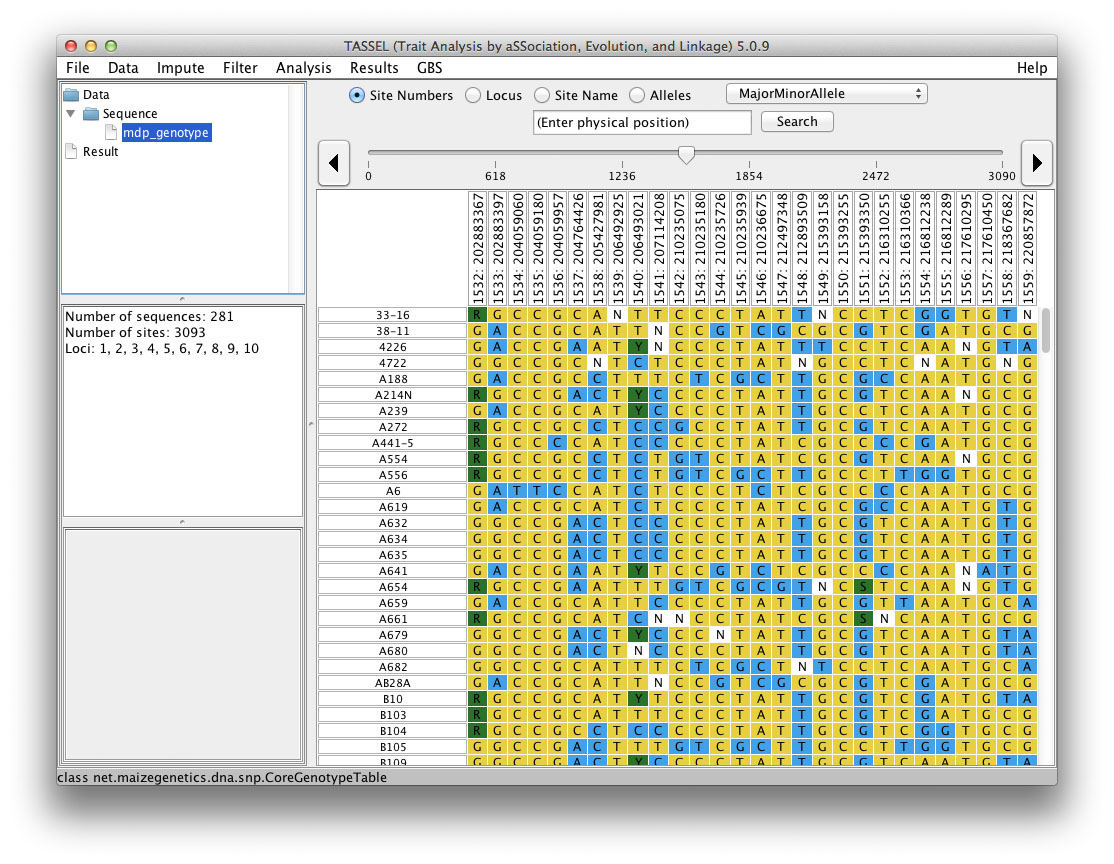
Alignment Viewer
|
TASSEL Version 5.0 Standalone
Using Git - Recommended! |
|
Archived Versions of TASSEL |
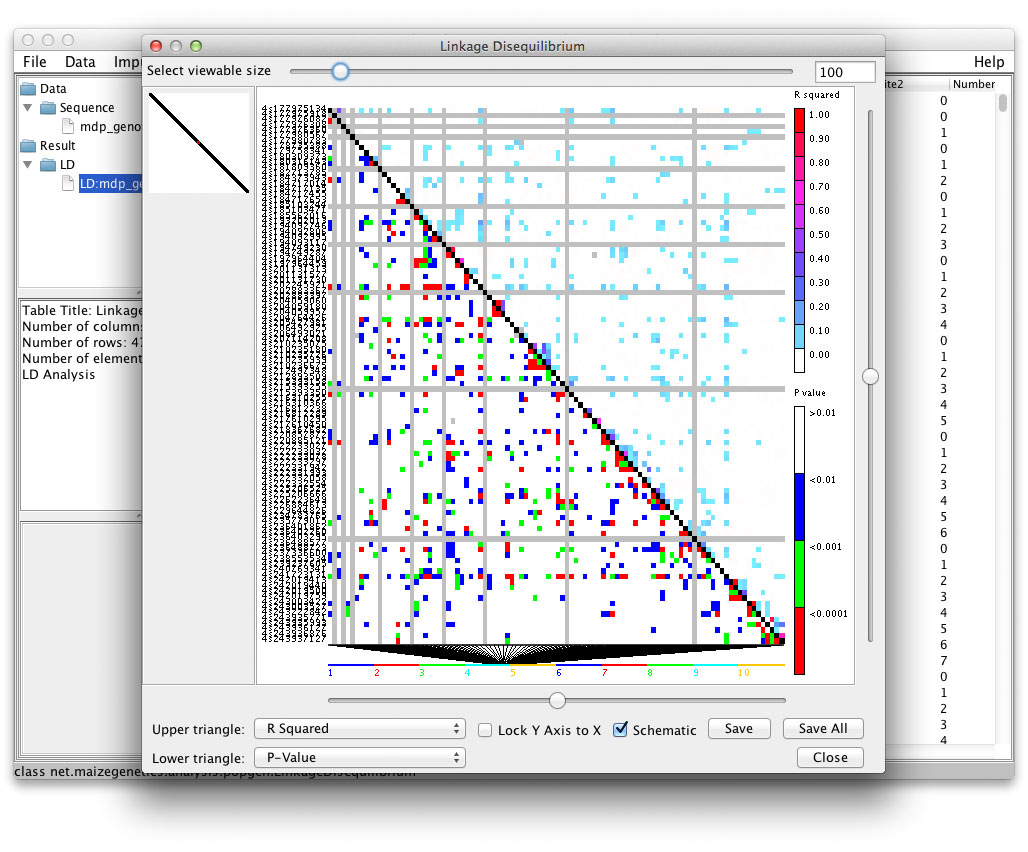
Linkage Disequilibrium Display
|
TASSEL DocumentationTassel 5.0 User's GuideR front-end for TASSEL (rtassel) Tassel 5.0 Youtube Tutorials Tassel Tutorial Data GBSv2 Test Data .tar .zip GBS Test Data Visit Tassel User Group Tassel 5.0 Change History Tassel 5.0 Wiki GNU Lesser General Public License |
|
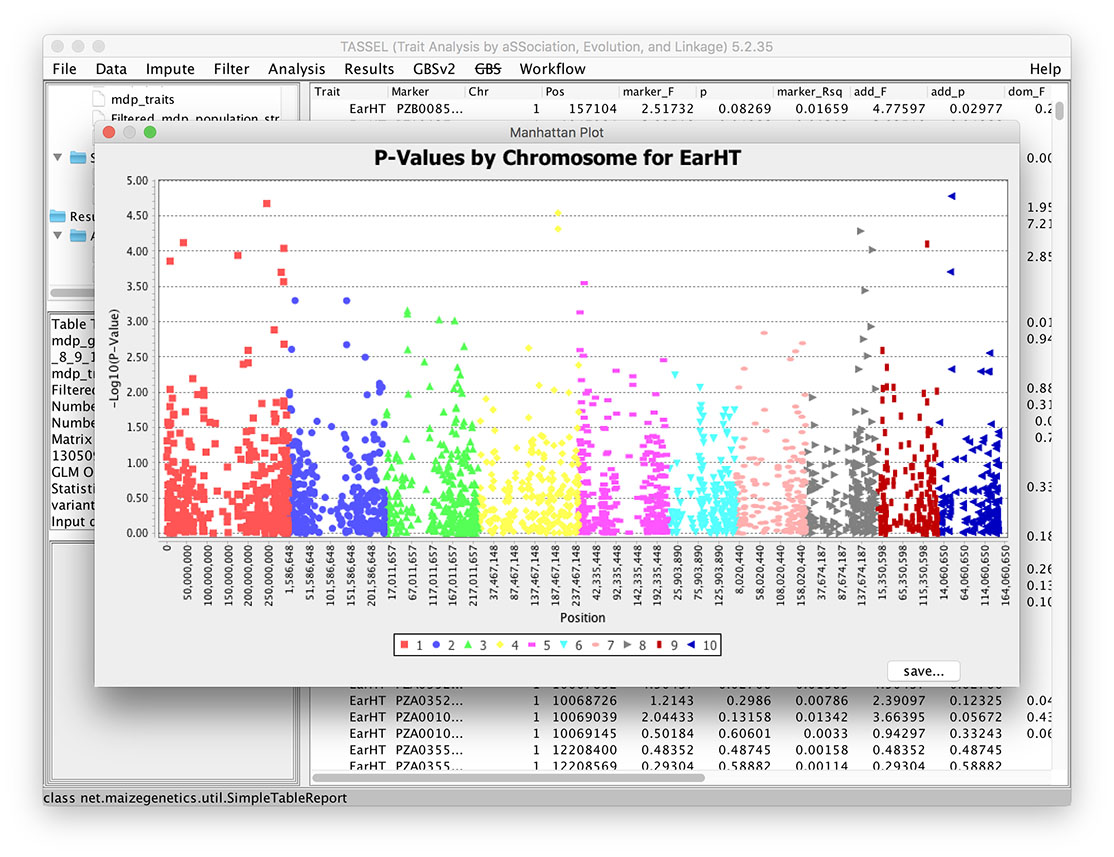
Manhattan Plot
|
|
TASSEL Pipeline DocumentationTassel 5.0 Pipeline DocumentTassel 5.0 GBS Pipeline v2 Document Tassel Pan-genome Atlas (PanA) Pipeline Tassel 5.0 Pipeline Tutorial |
|
TASSEL development has been generously supported by the USDA-ARS and the NSF, as well as free or reduced cost tools including:Bitbucket (Tassel 5.0)SourceForge (Tassel 3.0 and 4.0) IntelliJ IDEA   |
Genetic Distance Heat Map

|
Contacts
|
Phylogenetic Tree using Archaeopteryx

|
|
|
